boa mitogenomes
Assembling mitochondrial genomes for an entire genus of Caribbean boas
This is the research that I completed for my undergraduate thesis at UNC Asheville in the Reynolds Lab.
Read the full paper here.
Summary of Project

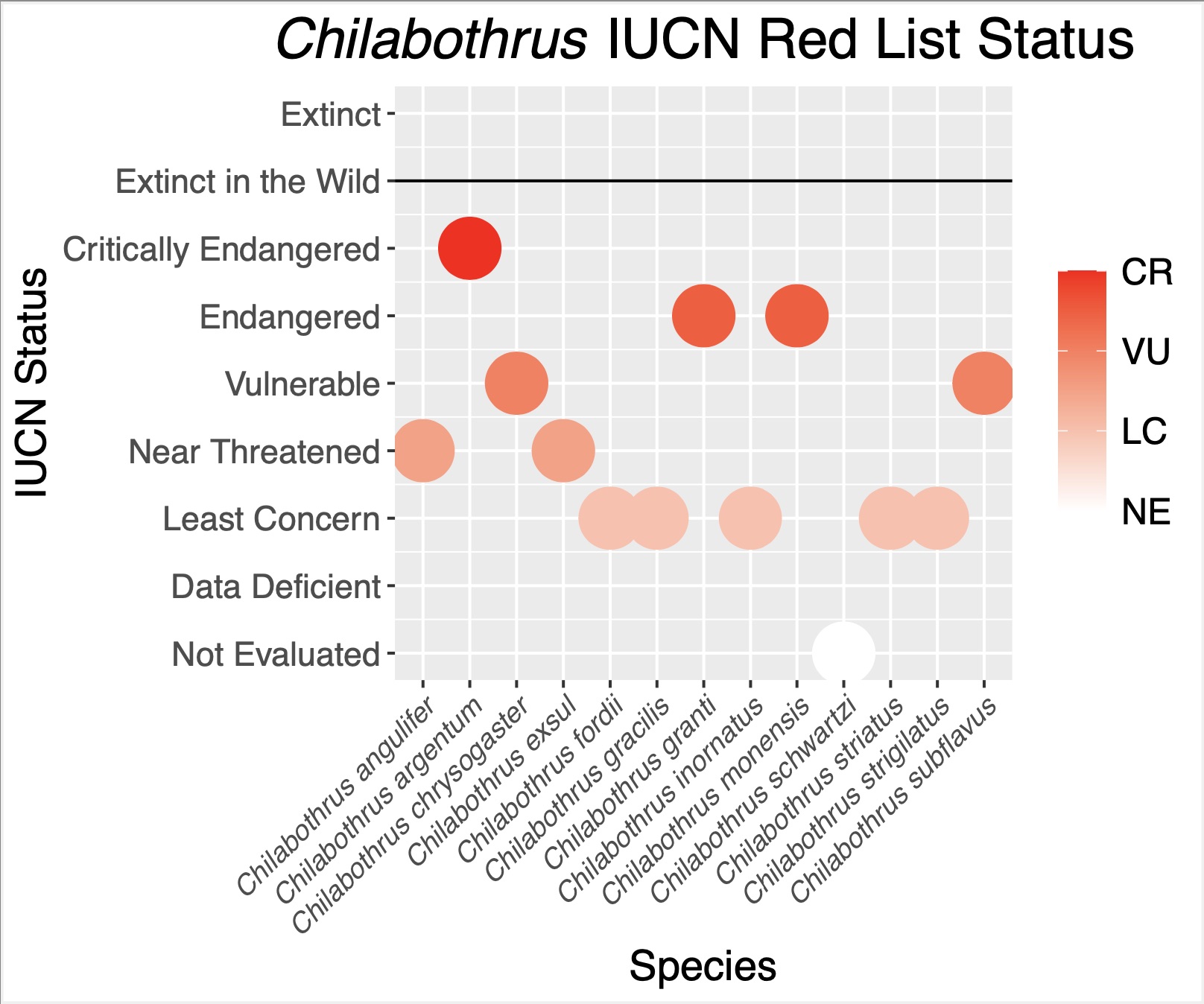
Chilabothrus is a completely insular genus of boid snakes occurring throughout the West Indies. The genus is composed of 14 species and nearly half of these are newly described or recently elevated1. Several species within this genus face serious conservation concerns such as introduced predators, anthropogenic habitat destruction, poaching for the pet trade, and low genetic diversity2,3,4,5. Two species are listed as endangered on the IUCN Red List, while the newly-discovered C. argentum is critically endangered and represented by as few as 135 remaining individuals (Figure 1).
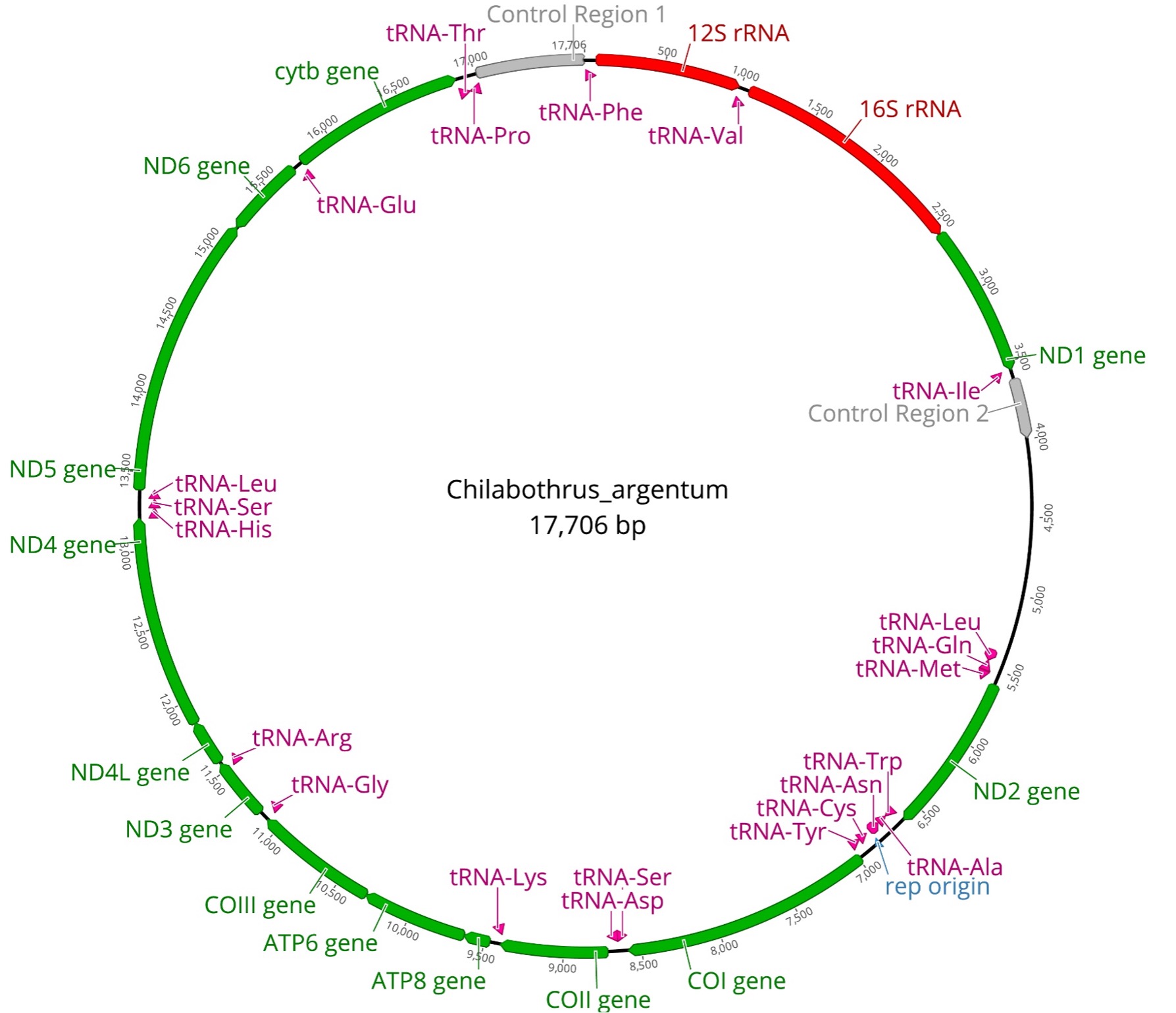
Mitochondrial DNA has long been a useful tool in phylogenetic studies6, with most studies focusing on sequencing one or two protein-coding regions. Since topologies tend to differ between studies using few versus many genes, we generated complete protein-coding mitogenomes for all Chilabothrus species (Figure 2). We used mitochondrial by-catch reads from ultraconserved elements enrichment and sequencing to assemble and annotate entire mitogenomes. These mitochondrial genomes are identical in composition and gene order to previously described boid snakes with 13 protein-coding genes, 22 tRNAs, 2 rRNAs, 2 control regions (CR1 and CR2), and an origin of the light strand replication (Figure 2)7.
Sequencing mitogenomes for threatened and endangered species is important in aiding conservation efforts by providing genomic resources. Providing these genetic resources for a threatened genus, such as Chilabothrus, will provide crucial information that can be used in studies focusing on their evolutionary relationships as well as those investigating population structure and genetic diversity.
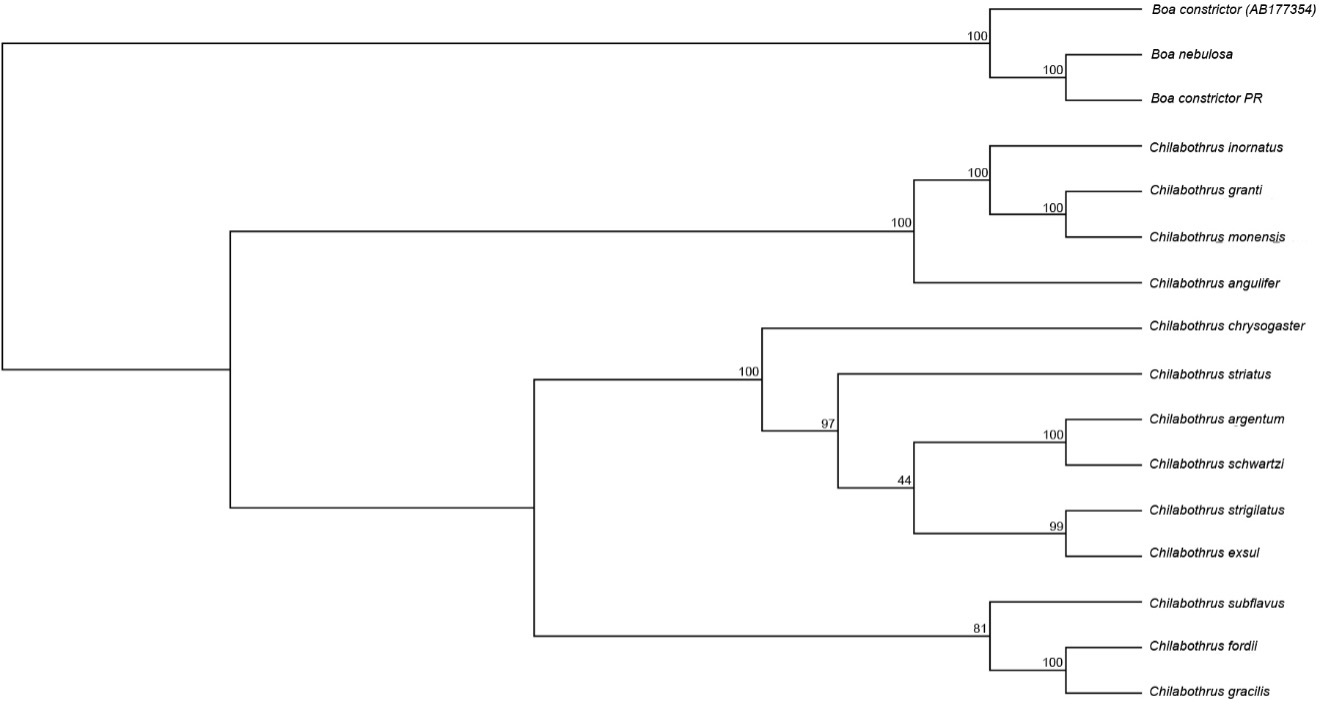
We generated a series of protein-coding molecular phylogenies of the entire genus were generated using Bayesian and Maximum Likelihood methods. The maximum-likelihood phylogeny produced a topology with strong support at most nodes (Figure 4). In concordance with previous studies8, we found that the newly-discovered C. argentum is sister to recently rediscovered C. schwartzi (BS=100; Figure 3). The topology shows that C. exsul and C. strigilatus are sister to one another with strong support (BS=99; Figure 3). This differs from previous topological arrangements using only the mitochondrial CYTB locus (~1100 bp) showing C. exsul more closely related to C. striatus and the clade containing C. argentum and C. schwartzi8. All other relationships agree with previously presented phylogenetic analyses.
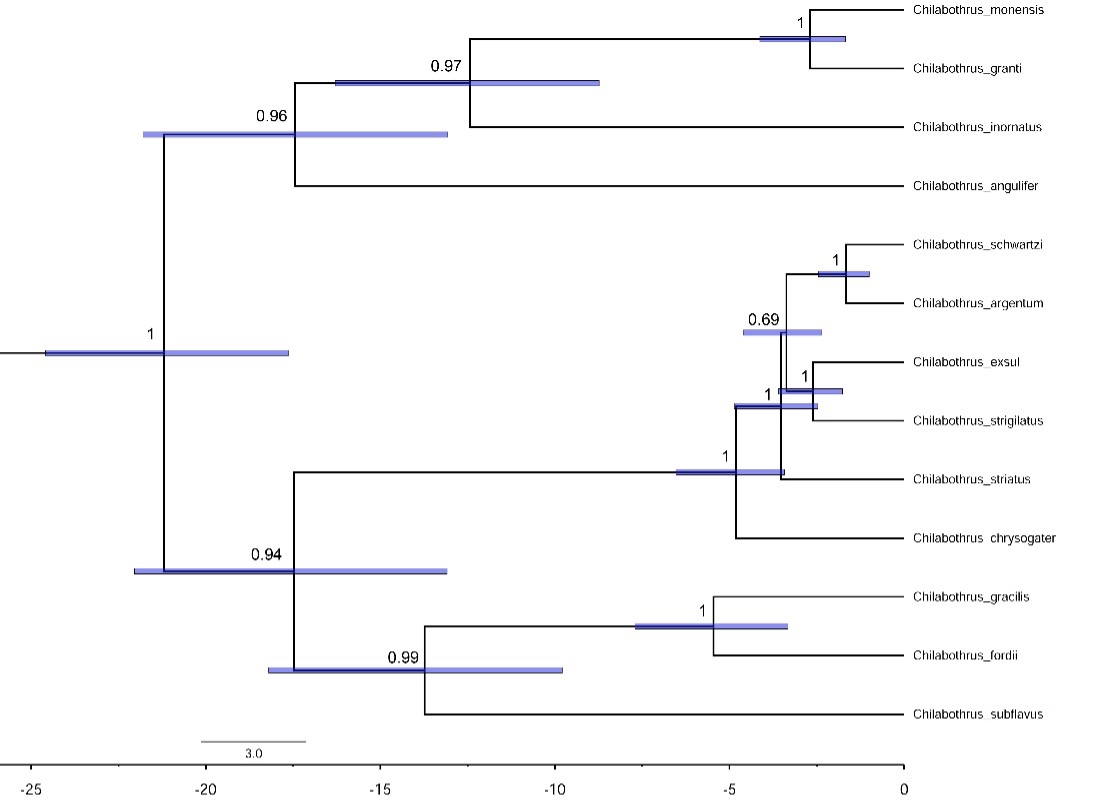
The maximum clade credibility tree produced the same topology as the maximum-likelihood tree (Figure 4). In addition to confirming the relationships among these taxa, this analysis presents divergence times of these species. We can use this analysis to estimate times of colonization events from Hispaniola. The first colonization event happened when C. chrysogaster colonized the Turks and Caicos around 5 million years ago. The second event happened around 3.5 to 4 million years ago when the clade containing C. schwartzi, C. argentum, C. exsul, and C. strigilatus colonized the Bahamas.
This is the first study to generate mitogenomes for an entire genus of boas. Being able to easily provide genomic resources for these threatened and endangered boa species is crucial for their conservation. Unlike previous studies that used one protein-coding region, the complete protein-coding mitogenomes have given a strongly supported topology for the Chilabothrus phylogeny. Our phylogenetic analyses support most previously published relationships of neotropical boids, only differing in topology in terms of the relationships of C. exsul, C. strigilatus, and C. striatus.
References
-
Reynolds RG, Niemiller ML, Hedges SB, Dornburg A, Puente-Rolón AR, Revell LJ. 2013. Molecular phylogeny and historical biogeography of West Indian boid snakes. Mol. 68:461-470. ↩
-
Reynolds RG, Puente-Rolón AR, Geneva AJ, Aviles-Rodriguez KJ, Herrmann NC. 2016. Discovery of a remarkable new boa from the Conception Island Bank, Bahamas. Breviora. 549:1-19. ↩
-
Reynolds RG, Gerber GP. 2012. Ecology and conservation of the Turks Island boa (Epicrates chrysogaster chrysogaster: Squamata: Boidae) on Big Ambergris Cay. J Herpetol. 46(4):578-586. ↩
-
Reynolds RG, Puente-Rolón AR, Platenberg R, Tyler RK, Tolson PJ, Revell LJ. 2015. Large divergence and low diversity suggest genetically informed conservation strategies for the endangered Virgin Islands Boa (Chilabothrus monensis) Glob Ecol Conserv. 3:487-502. ↩
-
Reynolds RG, Henderson RW, Díaz LM, Rodriguez-Cabrera TR, Puente-Rolón AR. In Press. Boas of the West Indies: Evolution, natural history, and conservation. Comstock Publishing Associates, Ithaca, NY. ↩
-
Raposo do Amaral F, Neves LG, Resende MFR, Jr, Mobili F, Miyaki CY, Pellegrino KCM, Biondo C. 2015. Ultraconserved element sequencing as a low-cost source of complete mitochondrial genomes and microsatellite markers in non-model amniotes. PLoS ONE. 10:e0138446. ↩
-
Dong S, Kumazawa Y. 2005. Complete mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 61:12–22. ↩
-
Reynolds RG, Puente-Rolón AR, Burgess JP, Baker BO. 2018. Rediscovery and a redescription of the Crooked-Acklins boa, Chilabothrus schwartzi (buden, 1975), comb. nov. Breviora. 558:1-16. ↩ ↩2